New Solutions to an Age-Old Problem
| "The key [to domestic breeding] is man's power of accumulative selection: nature gives successive variations; man adds them up in certain directions useful to him."
—Charles Darwin, |
The story of animal agriculture is a story of constant improvement. Ever since the first animal was domesticated, producers have repeatedly applied new knowledge to the perennial challenge of providing good and plentiful food to a hungry world.
One of the main ways in which people have worked to improve agriculture is through selective breeding—preferentially allowing animals with traits that make them most valuable to humans to produce young. While their less adequate counterparts became dinner, the hefty hog, the well producing milk cow, the broad-beamed steer, the prolific chicken became the parents of the next, slightly better generation. The result over many years is a spectrum of animal breeds and lines that are far more productive than their wild ancestors.
Hit or Miss
Though it sounds straightforward, animal breeders know all too well that selective breeding is in reality a frustratingly hit-or-miss proposition. That's because of the way in which the characteristics of offspring are determined.
 | For years, hog and other livestock producers have taken their champion animals and selectively bred them to improve the overall performance of their animal herds. |
Every animal has within the nuclei of its cells a set of molecules called chromosomes, half of which it inherited from its mother and the other half, from its father. Animals that belong to the same species generally have the same number and same kinds of chromosomes. However, even though the chromosomes look identical from one animal to the next, they contain minute and extremely significant molecular differences.
Those differences occur in the makeup of DNA, the long molecule that comprises the bulk of each chromosome and is duplicated and passed from generation to generation. DNA is formed from four kinds of smaller molecules known as nucleotides, linked together like beads on a necklace. For most of the length of each chromosome, the order of the nucleotides is the same in every member of a species. But along occasional stretches the pattern differs, creating inherited variations known as polymorphisms ("many forms").
Some polymorphisms occur in areas of the DNA that do little but get passed from one generation to another; these don't really affect what the organism is like. But not all DNA just sits there. In certain functional stretches of the chromosomes called genes, the nucleotides are used as a template to create proteins. These proteins, in turn, become the workers on the assembly lines that produce the features of an individual organism. Because different nucleotide sequences produce different proteins and different proteins create different assembly-line products, minute variations in the way nucleotides are ordered within genes are eventually translated into the remarkable kaleidoscope of diversity that we see every day in the organisms around us.
Every animal carries two of each type of chromosome—one received from its mother and the other from its father. That means that it also carries two genes for each trait. With few exceptions, each gene is either dominant, meaning that it is expressed no matter what the other gene is, or recessive, meaning that it is expressed only if the other gene for that trait is also recessive. Genes that are not expressed do not show up as a trait in the animal that carries them. However, they can be passed along to—and perhaps expressed in—that animal's offspring.
The hit-or-miss aspect enters into the picture in several ways. One has to do with the recessive genes—the ones that can be carried but may not be expressed. These genes can be transferred quietly from one generation to the next until an animal inherits one from each parent and the trait is expressed. When that happens, it may create big surprises for producers who have not paid adequate attention to the ancestry of their animals.
 "Mapping the swine genome will provide
researchers with the fundamental genetic knowledge necessary to
develop procedures and strategies that will allow genetic improvement
to occur at a more rapid rate and in a more predictable manner. The
more that is known about specific genes, where on the chromosome they
are located, how they are controlled, and how they function, the
better chance geneticists and swine breeders have of facilitating
favorable genetic change."
"Mapping the swine genome will provide
researchers with the fundamental genetic knowledge necessary to
develop procedures and strategies that will allow genetic improvement
to occur at a more rapid rate and in a more predictable manner. The
more that is known about specific genes, where on the chromosome they
are located, how they are controlled, and how they function, the
better chance geneticists and swine breeders have of facilitating
favorable genetic change."Richard Frahm, director National Animal Genome Research Program USDA- CSRS |
But even that isn't all. Some genes are expressed incompletely, meaning that they carry more information than actually shows in a given organism—information that can startlingly be expressed after it's been passed along to a new generation. And on top of that, in the process of selecting for desirable traits, producers often find that undesirable traits piggyback along from one generation to the next (witness the rise in porcine stress syndrome, or PSS, as producers select for lean genes using conventional breeding methods).
These genetic sources of uncertainty are further complicated by the fact that some important traits remain mysteries for much of an animal's life, resulting in long and expensive lead times for breeders working to improve their herds. For example, whether a pig inherits its sire's carcass quality will not be apparent until after it has been fed and housed for several months. In the same way, with conventional breeding a producer cannot tell if a young boar is carrying the genes that made its mother produce large litters until its offspring have pigs of their own.
 | Recent advances in molecular genetics are beginning to allow producers to breed animals for desirable traits by examining genetic material derived from a small blood or tissue sample. |
In a nutshell, the world of breeding is a chancy place, completely at the whims of the biological forces that rule heredity. Undesirable traits that an animal carries in its genotype (genetic makeup) but do not show in its phenotype (the way that genetic makeup is expressed as flesh and bone) can pop up unexpectedly in its offspring. Desirable traits may be inherited but not passed along, or inherited but not expressed. The whole process, in the end, makes every generation a bit like a freshly dealt poker hand, full of new combinations that could turn out to be big winners—or equally big losers. And just as in poker, though selective breeding involves skill, it is ruled by a discouraging amount of old-fashioned luck as well.
|
Landmarks of Animal Genetic
Improvement | ||
|---|---|---|
| 1866 | Law of Segregation | Gregor Mendel |
| 1908 | Genotype Frequencies | G. Hardy, W. Weinberg |
| 1910-40 | Heterosis | E. East, G. Shull, F. Keeble, C. Pellew |
| 1918 | Variance, Correlation
Between Relatives, Additive Genetic Variance—Heterozygosity, Linkage | R. Fisher |
| 1921 | Selection Within and Between Herds, Grading-Up, Inbreeding, Epistasis | S. Wright |
| 1932 | Fitness, Genetic Load and Cost of Selection | J.B.S. Haldane |
| 1942 | Index Selection | L. Hazel, J. Lush |
| 1949 | Recurrent and Reciprocal Recurrent Selection | R. Comstock, H. Robinson, P. Harvey |
| 1981 | Marker Assisted Selection | M. Soller |
| 1984 | Estimation of Variance Components, Prediction of Breeding Values, Single-Trait and Multiple-Trait Genetic Evaluations (BLUP) | C. Henderson |
| Mammalian
Genomes | |||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Just as it takes unfathomably large numbers of individual letters of the alphabet to create the compilation of information we know as an encyclopedia, so too it takes huge numbers of nucleotides to form a chromosome. And just as different encyclopedias have different numbers of volumes, different animal species have different numbers of chromosomes and other genetic marker characteristics: | |||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||
| The Music of
Life | |
|---|---|
| A picture of the relationship between chromosomes, genes and heredity is as close as your stereo. Take the CD you got for your birthday. It contains, in the unique order in which bumps and valleys are strung together along its surface, all of the information your CD player needs to know whether to play Beethoven or Bon Jovi. Yet to the eye, all CDs look the same. | Similarly, the difference between one chromosome and another lies in the order in which nucleotides appear. Just as a CD player "reads" bumps and valleys and converts the resulting information into sound, the code of the chromosome is translated by special chemicals into the incredible symphony of form and function we know as a living creature. |
A Closer Look
Enter genetic mapping.
Imagine being able to take a look at an animal's genetic material directly, to see whether the instructions for large litters or some other unexpressed or yet-to-be-expressed trait such as growth rate, reproduction or disease resistance lurks there. If producers could do that, they could make much more rapid and efficient decisions about which animals to breed and which animals to cull, saving huge amounts of time and money by increasing the rate of genetic progress in their herds.
Sound straightforward? It would be if chromosomes were the size of garden hoses and genes had iron-on labels describing the characteristics they code for. Unfortunately, that's not the case. Chromosomes are far tinier than can be seen by the naked eye, and individual genes—which themselves are only minute parts of the chromosome—are certainly not wearing signs publicizing what they do for a living. So until recently there has been no easy way to tell by examining genetic material whether the code for a certain trait is buried there.
Since the mid-20th century, however, scientific advances have made it possible to understand and work with the genome in a way that is revolutionizing agriculture, perhaps as much as the first shift from hunting to domestication. Beginning with the discovery of the structure of DNA in 1953, our understanding of how genetic material orchestrates the development of an entire plant or animal has snowballed, with knowledge leading to the development of new exploratory techniques, which in turn lead to further discoveries, and so on. Just within the last decade, an entire apartment building's worth of new doors have been opened with the commitment of research funds worldwide to creating a comprehensive map of the human genome. This massive cooperative effort has yielded extensive progress in laboratory technology, statistical science, mathematics, molecular genetics, computer capabilities and related areas.
 "We believe that in the future routine
genetic analysis will take place in breeding stock. . . . I think it
will be possible in the next five to seven
years to analyze at the genetic level to identify specific traits in a
breeding program."
"We believe that in the future routine
genetic analysis will take place in breeding stock. . . . I think it
will be possible in the next five to seven
years to analyze at the genetic level to identify specific traits in a
breeding program."Stephen Bates Group Leader, Agriculture Perkin-Elmer, Applied Biosystems Division |
In the early 1990s, animal geneticists proposed a concerted effort to apply this treasure trove of information and technology to improving food animal health and productivity through gene mapping. Efforts such as the USDA-funded National Animal Genome Research Program (NAGRP), the European Community's PiGMaP (mapping the pig genome) and BovMap (mapping the bovine genome) projects, and work by the U.S. Agricultural Research Service Meat Animal Research Center and Beltsville Area Research Center coalesced into a critical mass that today is on the verge of transforming sophisticated molecular genetics from an intriguing but largely cerebral pursuit into a results- oriented process for applying selective breeding at the level of the gene rather than of the whole animal.
Decisions about the application of genetic technology gleaned in the laboratory ultimately rest in the hands of producers and those they consult when making production decisions. The quality of these decisions, in turn, depends on an adequate knowledge of the layout of the playing field—hence, this primer. In the pages that follow, we describe current efforts to map the pig genome. We also discuss the various ways in which growing knowledge in this area already is helping to improve production, and present some visions for future benefit as we work to enhance our ability to ensure a high-quality food supply for our world.
| A Look at
the System |
|
|---|---|
|
The story of heredity is the story of a chemical symphony buried deep within the nucleus of the cell. When a pig is conceived, it receives a haploid ("half") set of 19 large molecules (18 autosomal, 1 sex), called chromosomes, from its dam (the female genetic parent) and a similar set from its sire (the male genetic parent). Together, these contributions form the diploid ("double") array of 38 chromosomes (19 pairs) that contain all of the information needed to transform a fertilized egg into a pig with its own set of individual characteristics. The alphabetic code used to spell out this remarkable compilation of information has only four letters, known as nucleotides and represented in biochemical shorthand as A, C, G, and T. When linked together in pairs in ladder-step fashion, nucleotides take on the well-known helical shape of DNA, the primary substance that comprises chromosomes. It is from special, functional stretches of DNA known as genes that the cellular machinery makes proteins, the basic building blocks of life. A gene may consist of fewer than a thousand nucleotides, or of hundreds of thousands. It may code for an entire protein or for just part of one. In the same way that digital variations determine the notes and pitches and hues that emanate from a compact disc, the order in which A, C, G, and T appear in a particular gene dictate the exact nature of the proteins under construction—and ultimately, the form and function of the symphony we know as a living organism. | 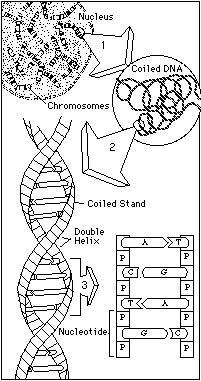 |